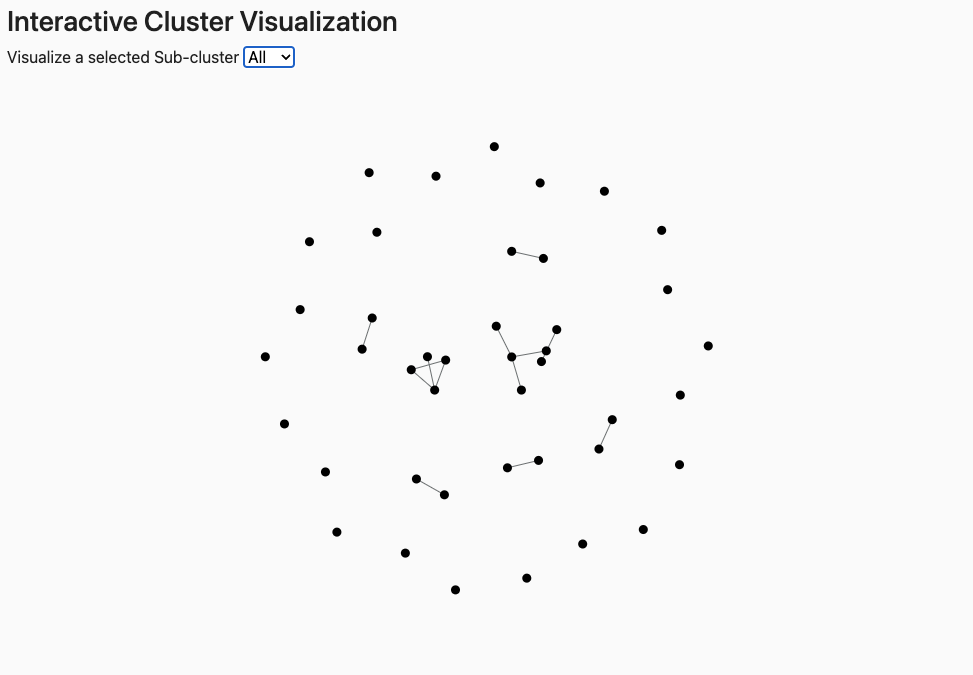
Cluster
The Cluster tool groups epitopes into clusters based on sequence identity. A cluster is defined as a group of sequences that have a sequence similarity greater than the minimum sequence identity threshold specified.
Parameter Selection
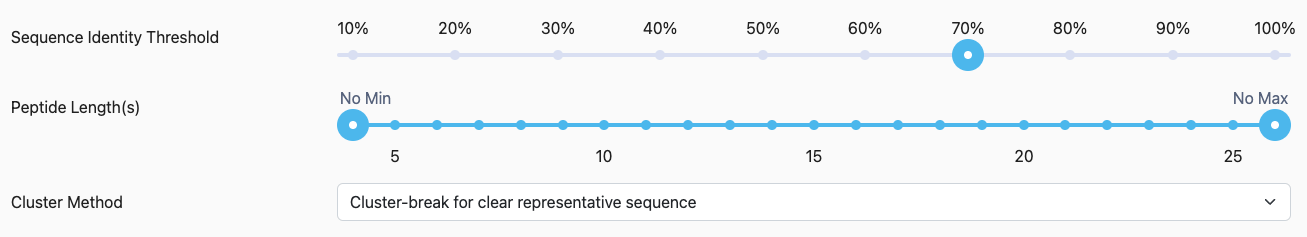
Sequence Identity Threshold
Select the sequence identity threshold at which you want to calculate epitope clusters.
Peptide Length(s)
Select the minimum and maximum length of peptides to consider for calculation
Cluster Method
Select one of the three approaches for clustering.
Clustering Methods
Before the different methods are described, we describe how the results are represented. The figure below represents a clustering result with 5 peptides (A, B, C, D and E) depicted as circles:
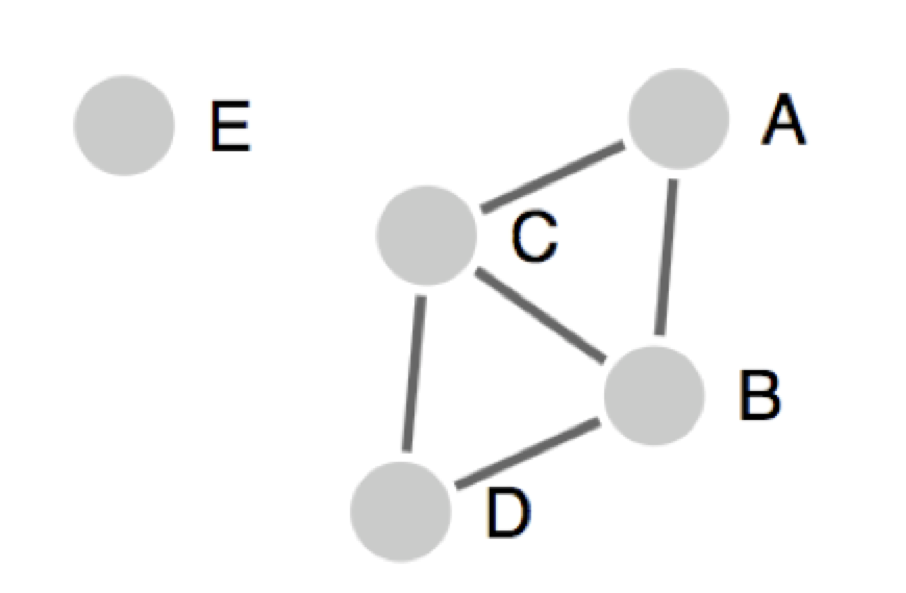
Lines (edges) connecting two peptides indicate identity above the specified threshold. Singletons are isolated peptides, like peptide E in the above figure, that don’t share sequence identity above the selected threshold with any other peptides in the given data.
1. All the connected peptides in clusters:
Here, all the peptides that are homologous to a certain pre-specified level are clustered together, say for example at the 70% level. In this case, any member of the cluster will be at least 70% homologous to at least one member of the cluster. However, the approach’s drawback is that members of the cluster might be related by levels of homology much lower than 70%. As a result, the cluster may not give a clear consensus sequence. Using this approach, in the above figure: A, B, C and D will make one cluster and E will be a singleton. Here, A and D are not directly connected, but these peptides will be a part of the same cluster.
2. Cluster-break for clear representative sequence (Recommended):
This is an extension of the first approach, where a cluster is broken down into subclusters at a point so that each subcluster can give a representative sequence. No peptide will be present in two clusters/sub-clusters. Using this approach, in the above figure: A, B, C and D will make one cluster, but these peptides can have different sub-clusters that have clean consensus sequences. E will be a singleton.
3. Fully interconnected clusters (cliques):
This is an alternative approach where all the peptides in a cluster are fully interconnected and share homology more than the given threshold. These fully interconnected clusters are called cliques. Here, one peptide can be a part of multiple cliques. Using this approach, in the above figure: A, B, and C will make one clique and B, C, and D will make a second. Note that B and C will be a part of both cliques in this case. E will be a singleton.
Results
The tabular output will include a column indicating the cluster and subcluster for each peptide. Subclusters beginning with the same digit before the decimal (e.g., 1.1, 1.2, 1.3) are derived from common parental clusters.
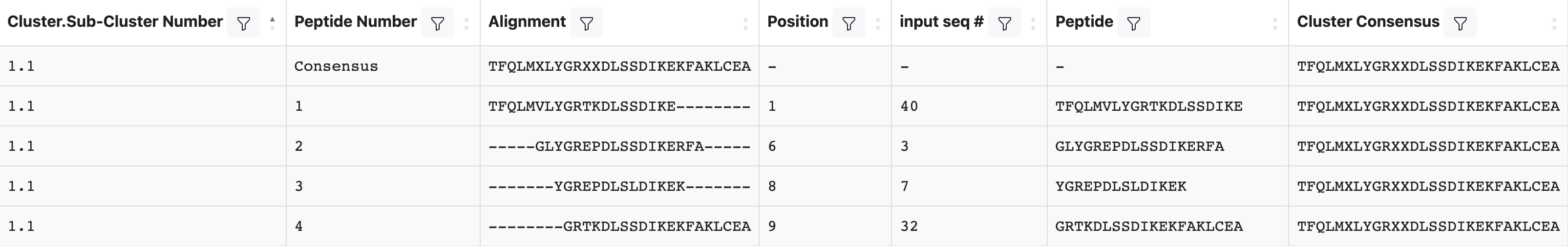
Cluster.Sub-Cluster Number
Cluster numbers are the sub-clusters broken down from common parental clusters. All the subclusters are given numbers with major clusters. Like 1.1, 1.2, 1.3 etc.
Peptide Number
The number of peptides in each cluster/sub-cluster/clique, where the first row in each will be the representative or consensus sequence. Numbers are assigned from the N to C terminus.
Alignment
The alignment of the peptide to the consensus.
Position
The starting position of the peptide when aligned to the consensus sequence.
Cluster Consensus
The consensus sequence of the cluster.
Cluster Visualization
With default settings, all the clusters/cliques will be shown, but the user may select individual clusters to visualize. The peptide sequences are displayed when nodes are moused over. Note that the visualization may be difficult to discern with large datasets.