Patient Harmonic-mean Best Rank (PHBR)
The Patient Harmonic-mean Best Rank (PHBR) tool takes in a list of neoepitopes and a set of HLA alleles for a single patient. Each peptide will be run through class I and/or class II binding predictions and scored according to the harmonic mean of their ranks across the panel of alleles. Lower PHBR scores indicate a higher likelihood of response to immunotherapy.
Input Format Types
Input neoepitopes
The PHBR tool takes a list of neoepitopes, including their mutation position(s), as input.
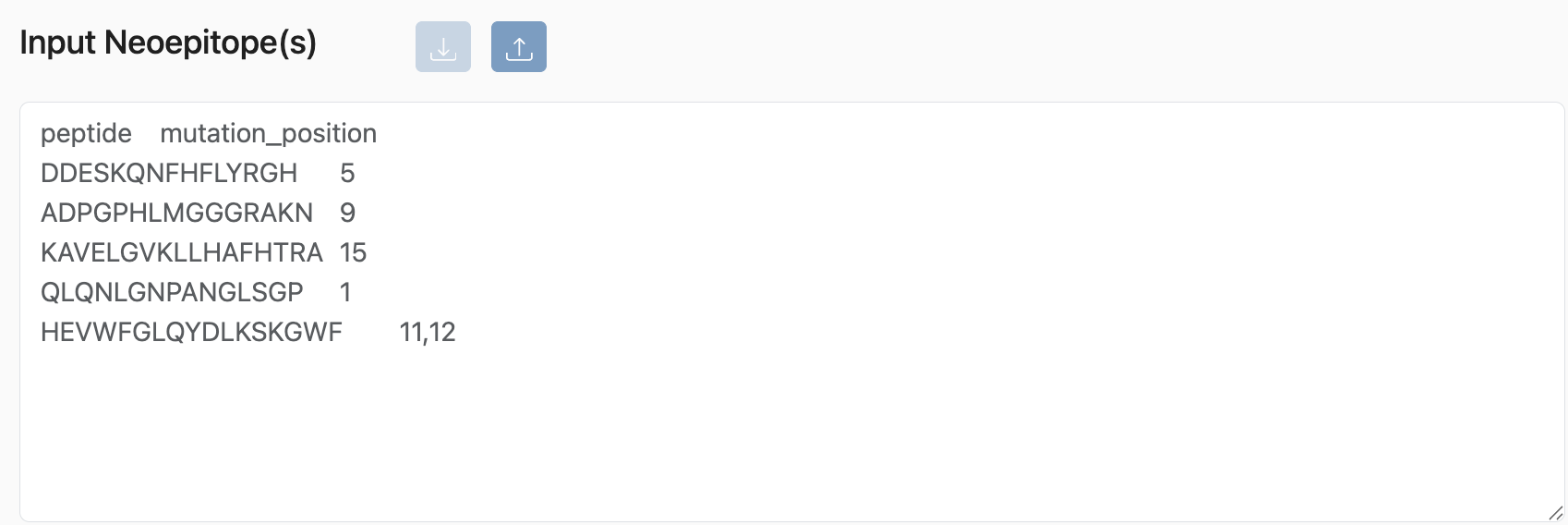
Example:
peptide mutation_postion
DDESKQNFHFLYRGH 5
ADPGPHLMGGGRAKN 9
KAVELGVKLLHAFHTRA 15
QLQNLGNPANGLSGP 1
HEVWFGLQYDLKSKGWF 11,12
The mutation positions are used to filter out predictions for sub-peptides that may not overlap with the mutations.
Paired Peptides
PHBR can also accept another input format where it takes mutated peptide and reference peptide. Note that the header must be present in the input.
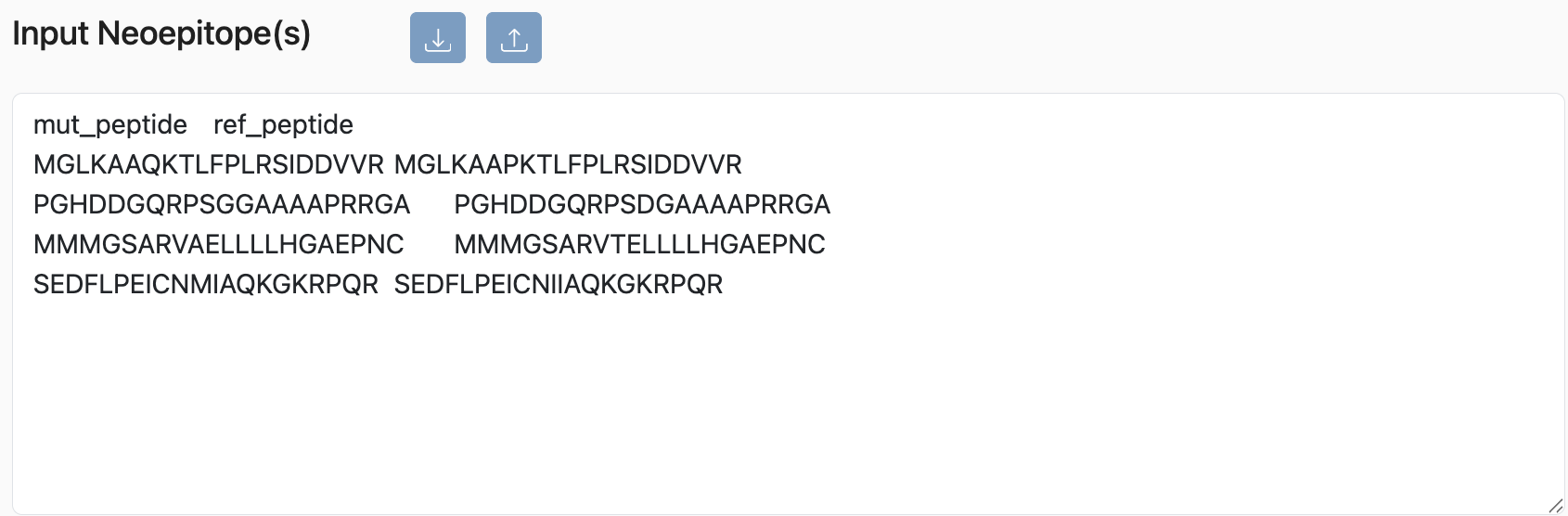
Example:
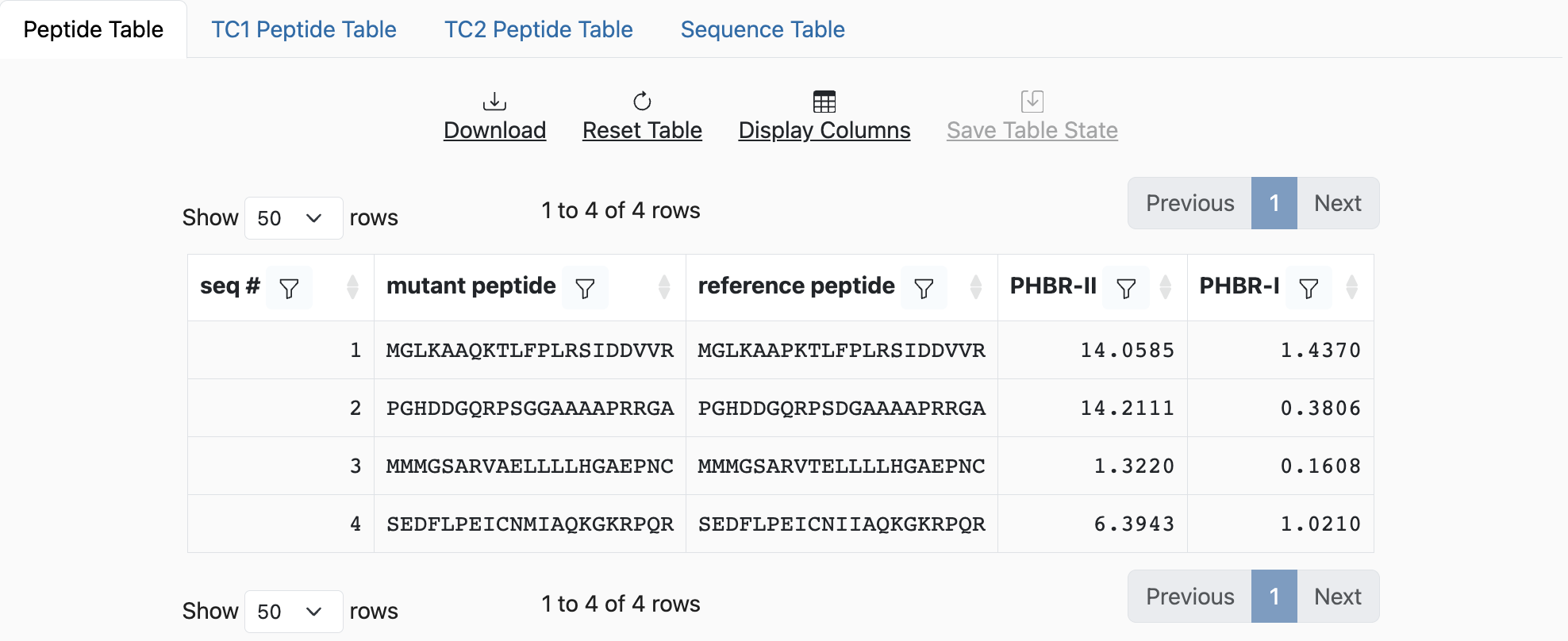
mut_peptide ref_peptide
MGLKAAQKTLFPLRSIDDVVR MGLKAAPKTLFPLRSIDDVVR
PGHDDGQRPSGGAAAAPRRGA PGHDDGQRPSDGAAAAPRRGA
MMMGSARVAELLLLHGAEPNC MMMGSARVTELLLLHGAEPNC
SEDFLPEICNMIAQKGKRPQR SEDFLPEICNIIAQKGKRPQR
Parameter Selection
The parameter selection for PHBR is straightforward and should look similar to below.
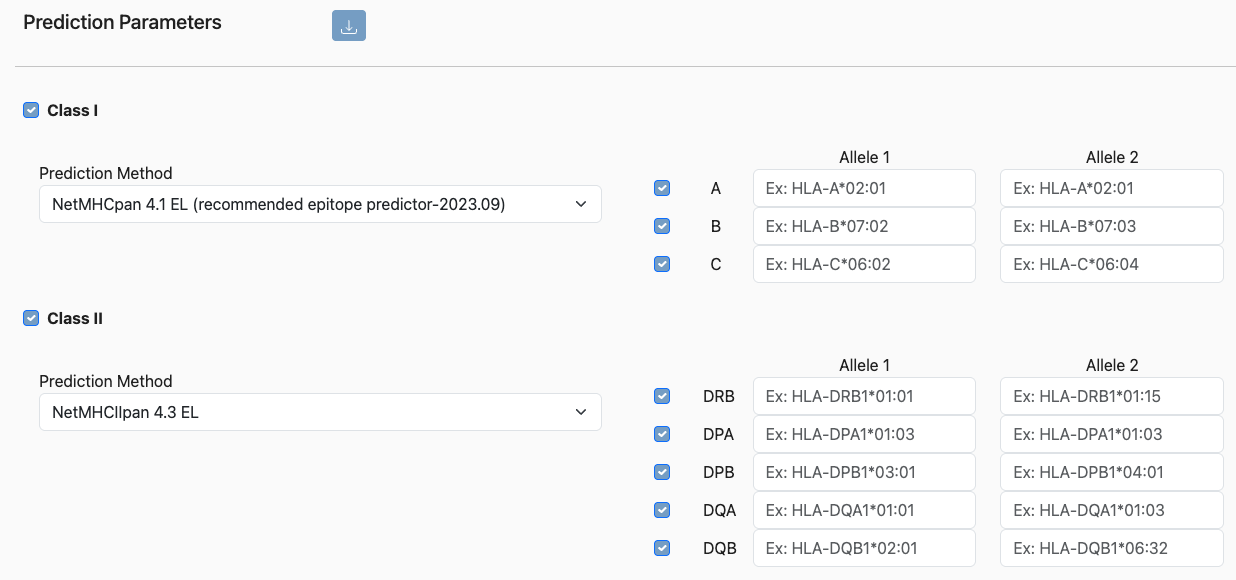
Class I and/or Class II
The user can opt to run either class I, class II, or both predictions on a set of peptides by checking/unchecking the appropriate boxes.
Binding/Elution method
Although we recommend using the default predictors, the user has the option to select alternative prediction methods from the Class I and Class II method dropdowns. Please refer to the Class I and Class II documentation for details on the individual methods.
Allele panel
In addition to the predictors, the alleles matching the HLA genotype of the individual should be selected. Note that class II alpha and beta chains are selected independently. When the prediction is submitted, binding predictions for all combinations of alpha and beta will be made.
Note
If an allele is left empty and the box is checked, it will be substituted with the example allele. To remove a locus from consideration, uncheck the box to the left of the locus name. If a locus is homozygous, denote that by entering the same allele for both Allele 1 and Allele 2.
Results
Output of this tool should include 1 row per input neopitope and up to two additional columns
with the PHBR-I and/or PHBR-II scores.
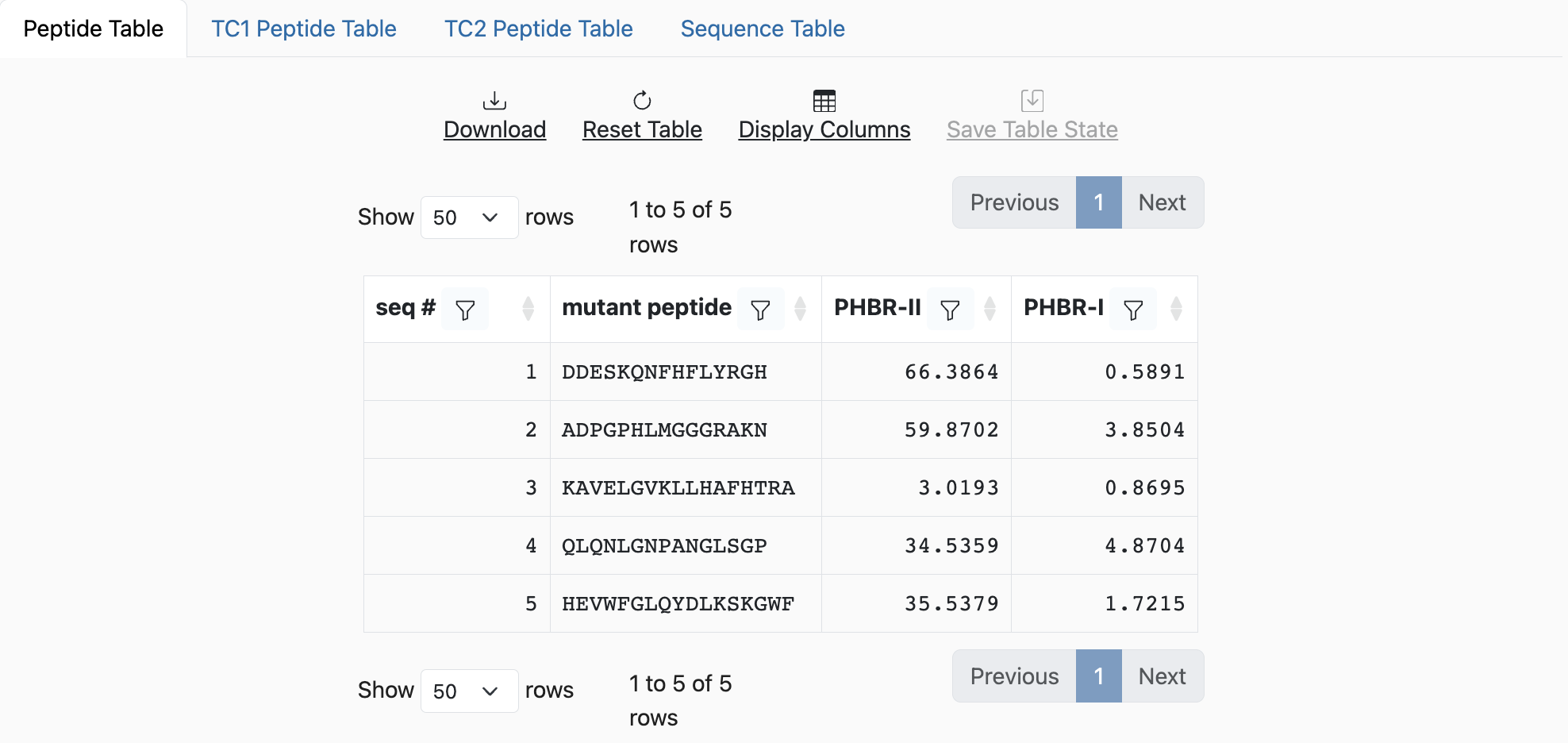
If the input was submitted as the paired peptides format, the output will show PHBR-I and/or PHBR-II scores, in addition to the paired peptides.
Interpretation
Lower PHBR scores (both I and II) indicate a higher likelihood of response to immunotherapy for this patient.