Peptide Variant Comparison
Peptide Variant Comparison
The peptide variant comparison (PVC) tool will take a paired list of wild-type and mutant peptides, and run predictors from the T cell class I application, or T cell class II application, and/or ICERFIRE.
Mode Selection
At the top of the form, mode selection is there to toggle between T cell Class I or T cell Class II.

Input Sequences
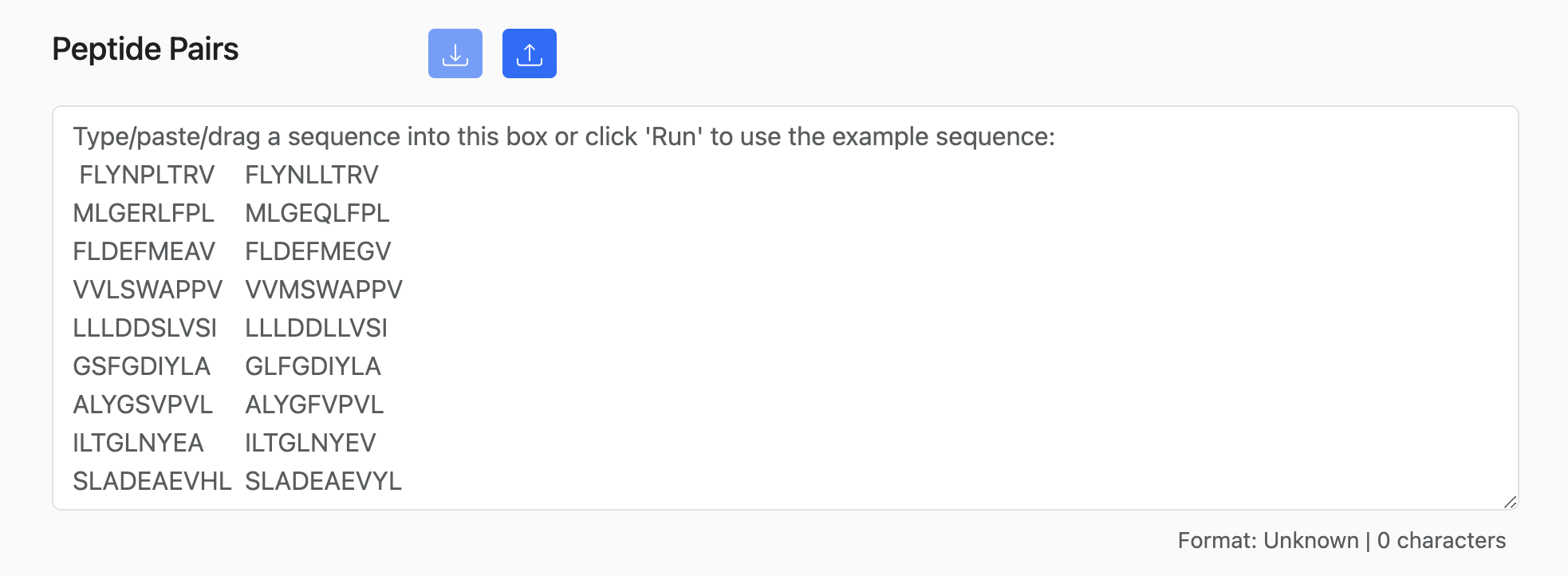 The PVC tool takes a list of paired wild-type and mutant peptide sequences as input. However, it can also take inputs with additional allele columns as well.
The PVC tool takes a list of paired wild-type and mutant peptide sequences as input. However, it can also take inputs with additional allele columns as well.
1. Peptide Pairs Only:
SVLDVGGAV,SVLDVGVAV
LLQVCERIPTI,LLQICERIPTI
PLLPALSPGS,PLLPALSPGL
IVSGVSQPDV,IVFGVSQPDV
SLLMWITQCF,SLLMWITQAL
2. Peptide Pairs and Alleles:
RRASKALLP,RRASKARLP,HLA-C*06:02
SSGFRNIAVK,SSGFCNIAVK,HLA-A*03:01
GKIPPSVQV,GIIPPSVQV,HLA-A*02:01
MASISSSLL,MASISSFLL,HLA-B*35:01
LQTIKDIASAI,LQTIKDIASPI,HLA-B*52:01
3. Peptide Pairs and TPMs (Only Compatible with ICERFIRE):
RRASKALLP,RRASKARLP,0.00199998
SSGFRNIAVK,SSGFCNIAVK,8.35072
GKIPPSVQV,GIIPPSVQV,105.084
MASISSSLL,MASISSFLL,6.201
LQTIKDIASAI,LQTIKDIASPI,36.9909
4. Peptide Pairs, Alleles, and TPMs (Only Compatible with ICERFIRE):
RRASKALLP,RRASKARLP,HLA-C*06:02,0.00199998
SSGFRNIAVK,SSGFCNIAVK,HLA-A*03:01,8.35072
GKIPPSVQV,GIIPPSVQV,HLA-A*02:01,105.084
MASISSSLL,MASISSFLL,HLA-B*35:01,6.201
LQTIKDIASAI,LQTIKDIASPI,HLA-B*52:01,36.9909
Notes
When alleles are specified in the input sequence box, these will override any allele selections made from the ‘MHC Allele’ selector.
The TPM column is only used for ICERFIRE. Note that ICERFIRE can use all four types of input formats.
The tool consider the first input column wild-type peptide and the second column mutant peptide. In the output, peptide A is wild type and peptide B is mutant, regardless of which method is chosen.
Delimiters
The paired wild-type and mutant peptides should be delimited by either
a tab or space.
The delimiter , or CSV file is not compatible
with the PVC tool.
Parameter Selection
The parameter selection UI is identical to T Cell Prediction - Class I
MHC alleles
Please refer to Getting Started > Prediction Parameters > MHC alleles for how MHC allele control section works.
Unsupported Alleles
If a target allele is unsearchable, it is most likely not curated yet.
Prediction models
Please refer to Getting Started > Prediction Parameters > Prediction models for how the prediction models control section works.
Available models for PVC Class I
As for the types of models that we provide, please reference Available Tools > T cell class I > MHC-binding predictions > Methods.MHC-I Binding/Elution
Class I Immunogenicity
ICERFIRE
Available models for PVC Class II
As for the types of models that we provide, please reference Available Tools > T cell class II > MHC-binding predictions > Methods.MHC-II Binding/Elution
Class II Immunogenicity
Results
Please refer to Getting Started > Prediction Parameters > Tabular Results for controls of the result table.
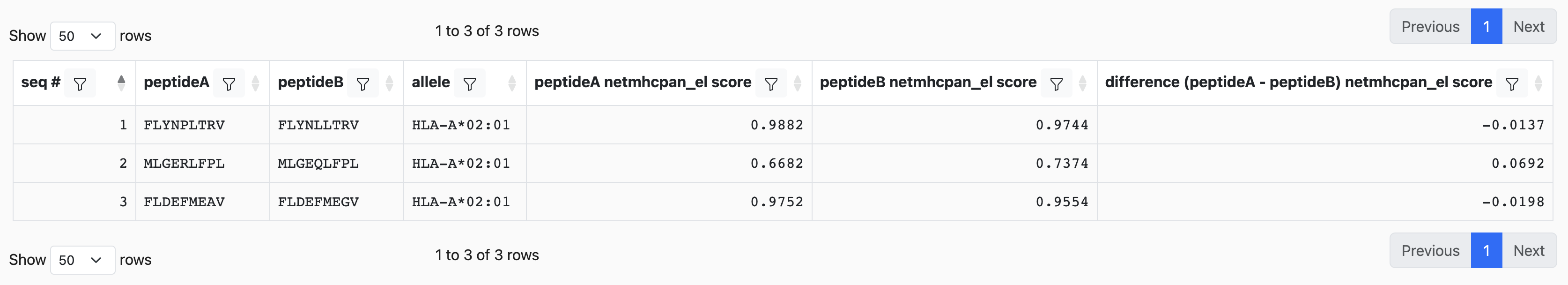
The result table will display prediction scores for each peptides, and also the difference between each prediction scores.
Scatter Plots
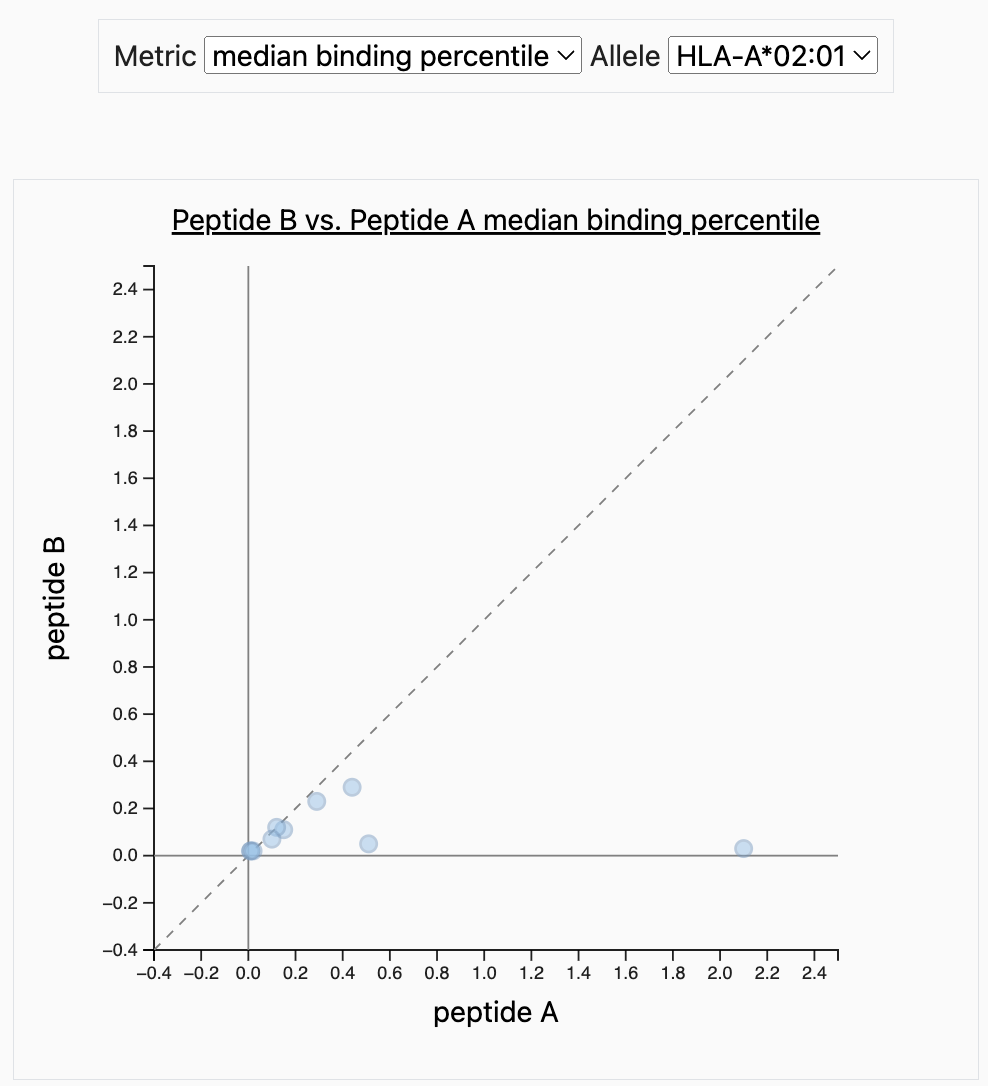
PVC Scatter Plot showing median binding percentiles.
Next to Peptide Table, there is Scatter Plots tab. This will display the results for each peptide on a graph. The default metric is set to median binding percentile.
Neo-Epitope immunogenicity prediction
ICERFIRE is an ensemble of Random Forest models. It predicts neo-epitope immunogenicity by identifying the best HLA-binding ICORE and uses self-similarity, a mutation score, and antigen expression.
Add the Neo-Epitope Immunogenicity using the Add Another Prediction button.
Parameter Selection
No additional parameters are needed.
Thresholds and Interpreting Scores
 The Icerfire Prediction score represents the immunogenicity of neo-epitopes, with higher scores indicating greater immunogenicity.
The Icerfire Prediction score represents the immunogenicity of neo-epitopes, with higher scores indicating greater immunogenicity.